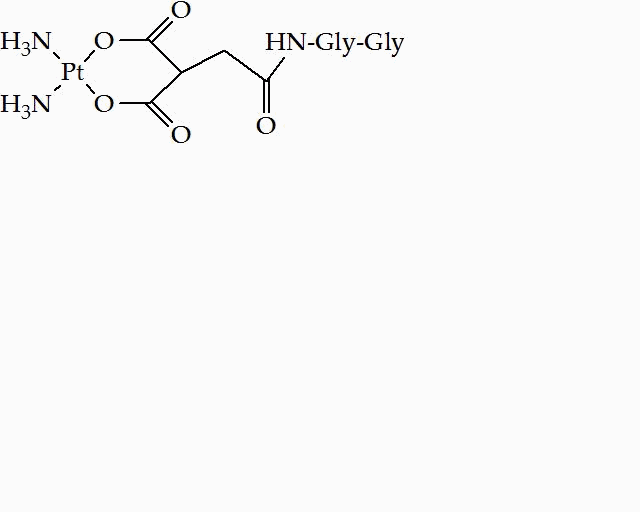|
Laboratory for Bioinorganic Materials and Biomolecules
Research Director
Dr. Nikos Katsaros, Tel: +30 210 6503645, +30 210 6503624, Fax: +30 210 6511766
Mrs.V. Kolyvaki
Dr. G. Psomas, Chemist, M.Sc. , Ph.D. Collaborating Scientists Dr. A. Scorilas, Chemist, M.Sc, Ph.D. e-mail: ascorilas@biol.uoa.grDr. A. Papakyriakou, Chemist, M.Sc, Ph.D. e-mail: thpap@chem.demokritos.gr
Mr. I. Bratsos, Chemist, M.Sc. e-mail: bratsos@chem.demokritos.grMrs. M. Katsarou, Chemist, M.Sc. e-mail:mariak@chem.demokritos.gr Mr. P. Christofis, Chemist, M.Sc. e-mail:petros_x@chem.demokritos.gr Mr. K. Floros, Chemist, M.Sc. Mrs. E. Efthimiadou e-mail: elefth@chem.demokritos.gr
Research Activities A. Synthesis and Characterization of new complexes of biological interest Our laboratory focuses on the synthesis and characterization of metal ion complexes with potential cytostatic agents as ligands. Attention has been attracted on platinum (II) and ruthenium(II) complexes. Structural characterization of metal ion complexes with anticancer drugs is performed using NMR techniques, biochemical methods molecular dynamics and X-ray. Studies on the effect of metal ion complexes at the structure and conformation of DNA are also being performed.
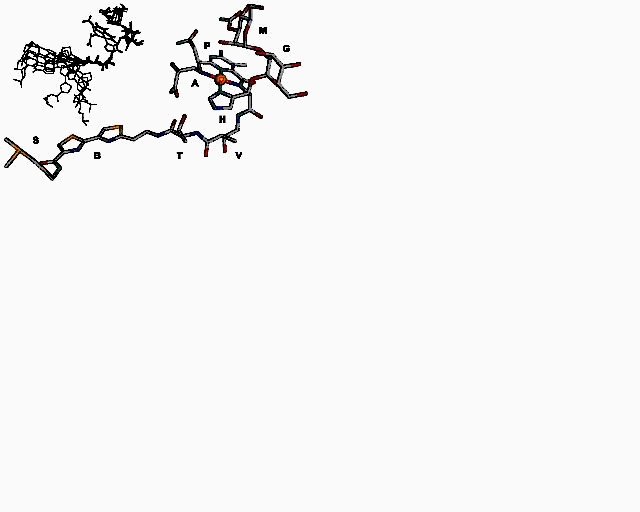
Structural Studies of the Metal Ion Complexes of the Anticancer Drug Bleomycin and Their Interaction with DNA Using Nuclear Magnetic Resonance in Combination With Molecular Dynamics Simulations The solution structure of three complexes of the anticancer drug bleomycin (BLM) has been carried out using high-field NMR methods in combination with molecular dynamics simulations. The stability of Ga(III)–BLM has allowed us to assign all the exchangeable amine and amide protons for the first time, which in turn was proven to be a valuable piece of information for its solution structure. NMR data were used as constraints in conjunction with simulated annealing molecular dynamics calculations. The NMR structure revealed that Ga(III)–BLM shares a similar structure with Co(III)–BLM complexes. By virtue of the fact that Ga(III) adducts are used as a probe for the biologically relevant Fe(III) compounds, their binding mode with BLM is proposed to be similar. Additionally, its interaction with the self-complementary oligonucleotide d(CCAGGCCTGG)2 was investigated by means of NMR. Our data, in comparison with those obtained for Co(III)–BLM, indicate that the drug binds strongly via intercalation of the bithiazole moiety between the central d(-CC-) step and that the pyrimidine ring of BLM might be involved in the formation of a base-triple-like interaction with the adjacent (G•C) base pair. This was the basis for the sequence specificity of DNA cleavage mediated by BLM at d(G-C) and d(C-T) sequences. Employing the same methods, we have also determined the solution structure of In(III)–BLM, which is successfully applied as radioactive imaging agent. Studies were performed at acidic solutions as well, with the aim to investigate whether structural changes could occur upon preparation of the kit for clinical use. The results indicate that In(III)–BLM has a similar structure with that of Ga(III)–BLM, which is not dependent on the acidity. Finally, the interaction of Pd(II) ions with BLM was studied in solutions with high ionic strength . In this way, it was found that one major species is present, which was characterized using NMR and molecular modeling. The force field AMBER(96) has been used throughout the molecular dynamics calculations, which was carefully parameterized in order to accurately represent the above compounds. All atom-centered point charges were derived from the electrostatic potential of BLM molecular fragments, which was calculated using the ab initio program GAMESS.
Structure of Pd(II)·bleomycin A2 Synthesis of carboplatin analog via Solid-Phase approach It is well known that the tumor resistance to platinum drugs is derived from the removal of cisplatin- and carboplatin-induced DNA adducts. A number of platinum complexes, which interact with DNA in a manner distinct from that of the initial drugs, have been designed and synthesized in order to overcome the tumor resistance. Amino acid residues, peptides, polyamides and oligonucleotides have been employed as site-specific DNA-interacting elements conjugated to metal complexes. Unfortunately, only few of these compounds were successful. Thus, the development of a methodology which allows a rapid synthesis of a wide variety of platinum(II) complexes is necessary. The synthesis of platinum complexes via solid phase approach is a very convenient and promising method. In the present work, a platinum(II) complex, a carboplatin analog, was synthesized. The results of earlier works from this laboratory in addition with the results of this project underline the fact that a number of peptide-containing platinum (II) complexes with potential antitumor activity can be synthesized.
Solid phase synthesis of the carboplatin analog
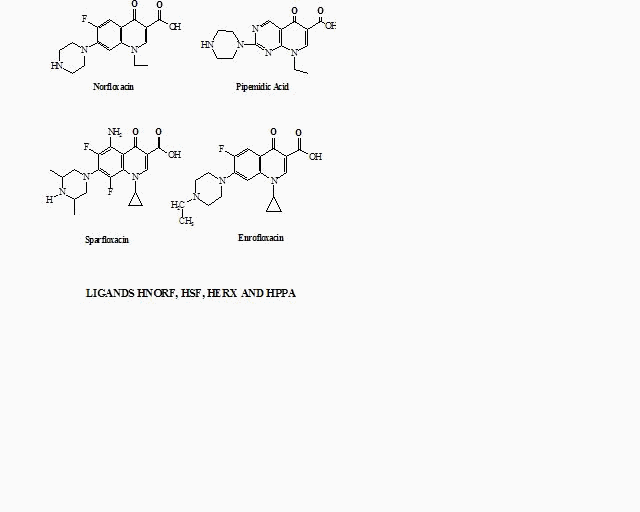
Synthesis and study of quinolone complexes-Structure and bioactivity Quinolones, a commonly used term for the quinolonecarboxylic acids or 4-quinolones, are a group of synthetic antibacterial agents widely used in clinical practice and act effectively by inhibiting DNA replication1. Norfloxacin, (1-Ethyl-6-fluoro-1,4-dihydro-4-oxo-7-(1-piperazi-nyl)-3-quinolinecarboxylic acid = HNorf),Sparfloxacin(5-amino-1-cyclopropyl-7-(cis-3,5-dimethyl-1-piperazinyl)-6,8-difluoro-1,4-dihydro-4-oxo-3-quinolinecarboxylic acid = HSf) and Enrofloxacin (1-cyclo-propyl-7-(4-ethyl-1-piperazinyl)-6-fluoro-1,4-dihydro-4-oxo-3-quinolone-carboxylic acid = HErx),are a typical fluoroquinolones used to treat gram-negative urinary tract infections. Pipedimic acid, (8-Ethyl-5,8-dihydro-5-oxo-2-(1-piperazi-nyl)pyrido(2,3-d)pyrimidine-6-carboxylic acid =HPPA) is a quinolone used to treat not only urinary tract infections, but also severe DNA-damages. Quinolones can act as bidentate ligands through the quinolone oxygen and one carboxylate oxygen atom2. In the literature, it has been reported that copper complexes with drugs are much more active in the presence of a nitrogen donor heterocyclic ligand, such as 2,2’-bipyridine (bipy), 1,10-phenanthroline (phen), 2,2’-dipyridylamine (bipyam)3-4. The interaction of Cu(II) with the quinolones norfloxacin and pipemidic acid in the presence or not of a N-donor heterocyclic ligand (bipy, phen, bipyam) has been studied in an attempt to examine the mode of binding and possible synergetic effects. In this contribution we report the synthesis and the characterization (with elemental analysis, IR, UV-Vis and EPR spectroscopy and C.D) of the mixed ligands copper complexes as well as the interaction of the complexes with DNA and their biological activity against microorganisms. The structures of the complexes Cu(Norf)(bipy)Cl and Cu(Norf)(phen)Cl (Figure 2) have been determined with X-ray diffraction.

B. Protein expression, structural and biological determination In collaboration with Magnetic Resonance Center (“CERM”, Sesto Fiorentino, Italy) several members of our group got involved with protein expression isolation and characterization using NMR spectroscopy. In more detail the proteins that were expressed and structurally characterized using high-filed NMR techniques are YPMQ and HAH1 protein (which implicate in copper homeostasis) and two domains of the metalloprotein calmodouline. Also, the C-terminal domain of a new protein, which involves in pre-mRNA splicing called SR-A1, is expressed and its interaction with known proteins is being examined.

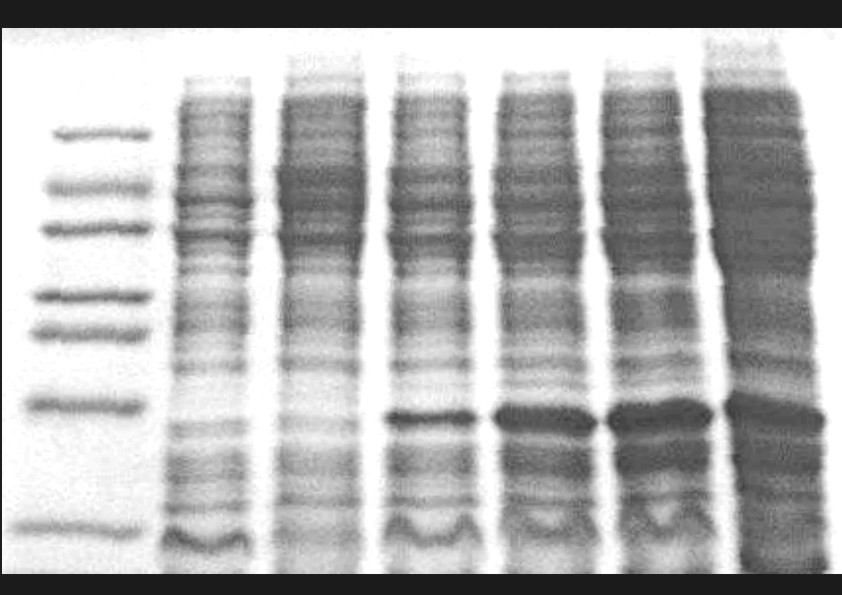
C. Gene expression in cell lines Cytotoxicity tests of potential cytostatic agents and their complexes in leukemia cell lines are performed, using known techniques (MTT, trypan blue). Induction of apoptosis by potential anticancer drugs is tested using DNA fragmentation technique (DNA laddering). Also, expression of the genes involved in splicing SR-A1, SRp20 and SC35, the apoptotic gene BCL2L12 and the genes BCL-2, Caspase 9, Caspase 3 Bax, Bcl-XL/S, Fas-Ligand and Fas in various leukemia cell lines under the effect of cytotoxic agents are being examined using molecular biology techniques.
Research Grants
Participating in Cost European Cooperation in the Field of Scientific and Technical Research
International Collaborations
Research Facilities
The main research facilities available at Laboratory include: Human leukemia cell lines (U937, K562, HL60) routinely maintained and stored at the laboratory (incubator 37˚C, 5% CO2), Laminar flow cabinet, optical and reverse microscope, PCR, autoclave, UV transilluminator, UV spectrophotometer. Also, the instruments that are available in the Institute of Physical Chemistry include NMR spectrophotometer, Cyclic Dichroism, EPR, X-ray crystallography and elemental analysis.
|